遺伝子発現機構学研究部門(前田教授)の嶋田誠講師の研究成果が国際誌(Molecular Phylogenetics and Evolution)に発表されました。
概要
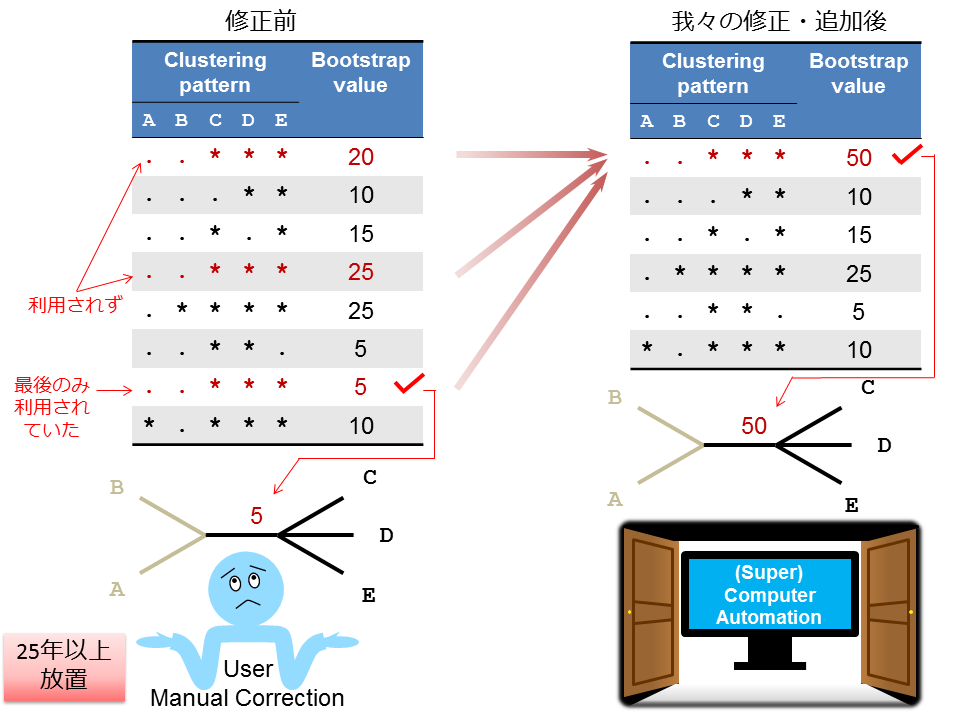
分子進化解析ソフトウエアパッケージの先駆けとして世界中で愛用され続けている、PHYLIP packageにおいて、25年以上もの間放置されていた、不具合の原因を突き止め修正したうえで、自動化へ向けた補強アドイン・プログラムとともに公開しました。
A modification of the PHYLIP program: A solution for the redundant cluster problem, and an implementation of an automatic bootstrapping on trees inferred from original data.
Makoto K. Shimada and Tsunetoshi Nishida
Molecular Phylogenetics and Evolution 109: 409-414. DOI:10.1016/j.ympev.2017.02.012
A modification of the PHYLIP program: A solution for the redundant cluster problem, and an implementation of an automatic bootstrapping on trees inferred from original data.
Makoto K. Shimada and Tsunetoshi Nishida
Molecular Phylogenetics and Evolution 109: 409-414. DOI:10.1016/j.ympev.2017.02.012
Highlights
- We corrected a software bug of PHYLIP, the evolutionary study package with the longest history.
- The bug had been left for more than 25 years.
- This correction solves the redundancy in bootstrapping that cause incorrect inference of consensus tree.
- We build an add-on program of PHYLIP that infers a tree from original data with the bootstrap values.
- We build an bash script that enable automatic tree construction from unaligned sequence data.
Abstract
Felsenstein’s PHYLIP package of molecular phylogeny tools has been used globally since 1980. The programs are receiving renewed attention because of their character-based user interface, which has the advantage of being scriptable for use with large-scale data studies based on super-computers or massively parallel computing clusters. However, occasionally we found, the PHYLIP Consense program output text file displays two or more divided bootstrap values for the same cluster in its result table, and when this happens the output Newick tree file incorrectly assigns only the last value to that cluster that disturbs correct estimation of a consensus tree. We ascertained the cause of this aberrant behavior in the bootstrapping calculation. Our rewrite of the Consense program source code outputs bootstrap values, without redundancy, in its result table, and a Newick tree file with appropriate, corresponding bootstrap values. Furthermore, we developed an add-on program and shell script, add_bootstrap.pl and fasta2tre_bs.bsh, to generate a Newick tree containing the topology and branch lengths inferred from the original data along with valid bootstrap values, and to actualize the automated inference of a phylogenetic tree containing the originally inferred topology and branch lengths with bootstrap values, from multiple unaligned sequences, respectively. These programs can be downloaded at: https://github.com/ShimadaMK/PHYLIP_enhance/.
問い合わせ先
| 研究に関すること | 報道に関すること |
| 研究者氏名 嶋田 誠(シマダ マコト) 藤田保健衛生大学 総合医科学研究所 遺伝子発現機構学研究部門 講師 〒 470−1192 愛知県豊明市沓掛町田楽ヶ窪 1-98 Tel:0562-93-9380 Fax:0562-93-8834 E-mail: mshimada@fujita-hu.ac.jp |
学校法人 藤田学園 法人本部 広報部学園広報課 〒 470−1192 愛知県豊明市沓掛町田楽ヶ窪 1-98 Tel:0562-93-2492 Fax:0562-93-4597 E-mail: koho-pr@fujita-hu.ac.jp |
| お手数ですが、スパムメール対策のため上記メールアドレス中の全角@を半角@に置き換えてご入力ください。 | |